Project Wonderland at the Chemistry Department, UWI-Mona
Educators are increasingly looking to technology to support
teaching and learning. The Open Source Project Wonderland
creates a virtual environment that was
originally implemented to support business collaboration, in
particular for the staff at Sun Microsystems, where on any given day many were
telecommuting or absent from office yet needed to actively
participate in meetings, etc. Following the takeover of Sun by Oracle
the project was forked and the Open Wonderland Foundation
established to continue development on the project.
Wonderland promises a rich
interactive visualisation experience that can focus, stimulate
and motivate learning in a dynamic environment where the
participant's avatars share and manipulate the objects of study
in real time. The JAVA-based tool-kit supports desktop
applications like Open Office, web browsers, voice telephony and
the display of video and PDF files. Developers can incorporate
their own JAVA applications and extend the functionality to
create new worlds and new features within existing worlds.
At the Department of Chemistry at the Mona Campus of UWI, Jamaica
we have established several Wonderland servers and have been
testing the incorporation of Jmol (for molecular graphics) and
JSpecView (for spectra) within these on-line environments. The
results so far suggest that this will provide an excellent
teaching and learning environment since users can interact with
the displays and discuss these using a whiteboard or through the
built-in audio and telephony features.

(A meeting held in Wonderland 0.5)
Jmol and JSpecView
A major factor in our choosing Wonderland was that Java
applications can be incorporated into the world without any need
for code changes. Thus Jmol and JSpecView run
interactively in-world unchanged.
Molecular graphics and spectra can then be selected by clients
and manipulated and with the full audio capability, discussions
can be held detailing features being highlighted.
Since an Internet browser window can be opened within Wonderland,
it is possible to display applets like Jmol and JSpecView from
existing web pages.
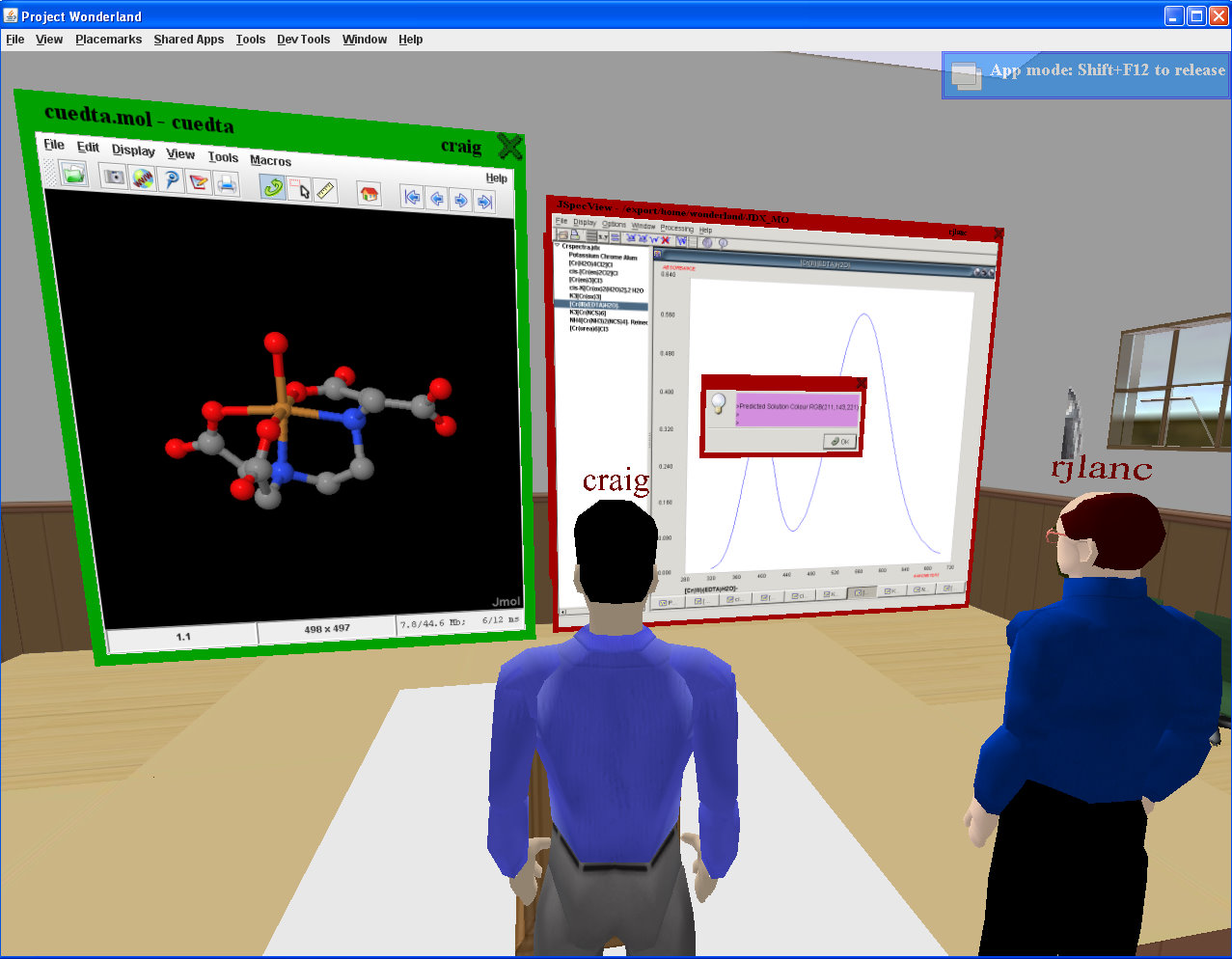
(A screen dump from Wonderland 0.4)
3D Models
We have developed an alternative approach by generating 3D
Collada models of molecular shapes and spectra.
For Jmol, we achieved this by exporting the shape as a VRML(WRL)
file, then using AccuTrans3D to convert this to .3ds and finally converting
to a Collada file using Google Sketchup.
See Cr(oxalate)3 ion - first isolated by Wilton Turner in London, 1830.
He was born in Clarendon, Jamaica in 1810 and was the brother of
Edward Turner - 1st Professor at the University of London (UCL 1828).
The models can be shown on web pages using the free 3D viewer
Once the model has loaded you can click in the right hand lower corner for Full Screen display
For JSpecView, we have done this by exporting as a SVG file (Inkscape version)
then using Blender to extrude the spectrum to give it some width, then
convert this to a .3ds file and again using Google Sketchup to create
the Collada file.
IR spectrum of p-chloroaniline
H NMR spectrum of vinylpyrrolidone
Accutrans3D is
shareware, Google
Sketchup is freeware and Blender and Inkscape are Open
Source.
The Google 3D
Warehouse is a handy resource for finding predrawn models for
use in Wonderland.
All of the models we have generated have been uploaded for
general use.
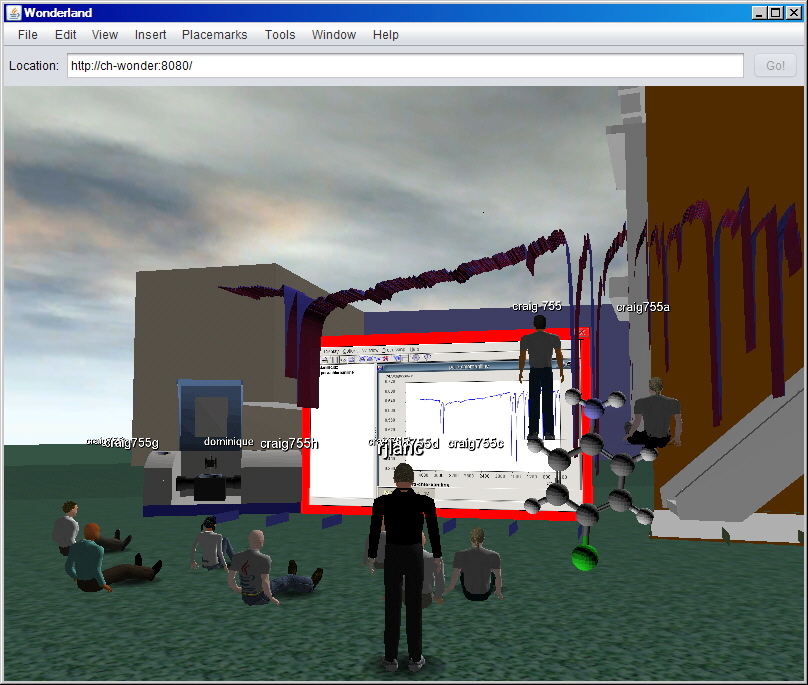
(A screen dump from Wonderland 0.5)
A virtual laboratory
We have started on a design of an online virtual laboratory
featuring various instruments including spectrometers (or chromatographs).
Eventually when the modules are fully functional we hope to be able to display
spectral data harvested from either local sources or from remote
databases.
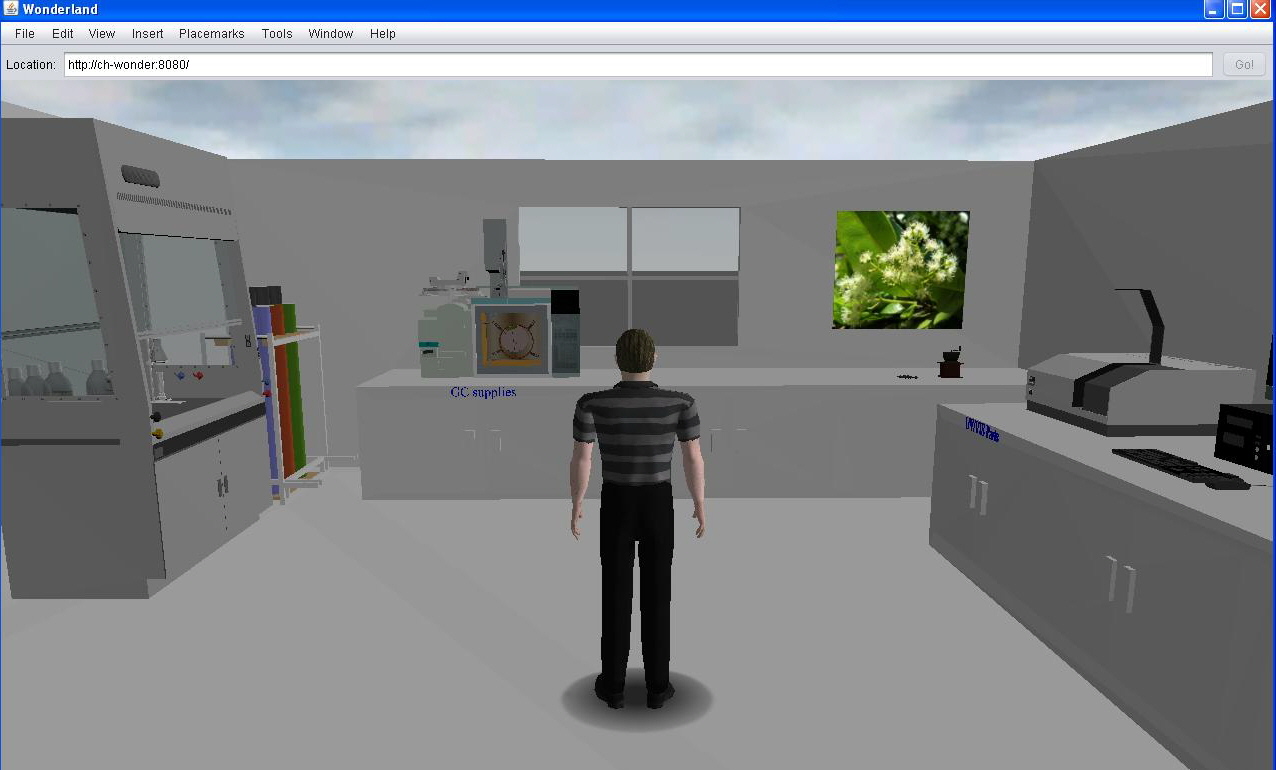
(A screen dump from Wonderland 0.5)
The first completed working module is of a Gouy Magnetic Susceptibility
setup. In our teaching laboratory the Gouy experiment can take upwards
of several hours to complete and with a class of over 100 students this
is a major timetabling problem. In the module, a large electro-magnet is shown
and the student is asked to select the voltage for generating the
magnet field, then choose an empty tube and weigh it with the
magnet switched Off then On, weigh a tube filled to a specified mark with water
then choose a calibrant (from 3 standards) and finally a sample
(from 6 different Chromium(III) complexes they might prepare in the Laboratory)
and repeat the weighings with the power Off then On.
An HUD shows the status and eventually gives the summary of all the recorded
data. This data can be copied to a text editor or spreadsheet and the
magnetic moment then calculated.
This module is part of an M.Phil project being undertaken by Mr Craig Walters.
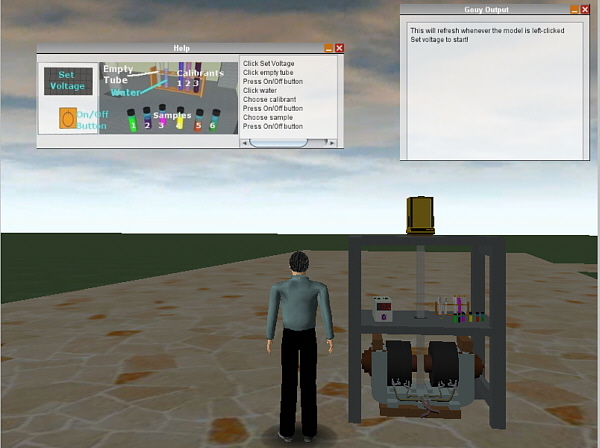
(A screen dump from Wonderland 0.5 showing the Gouy module)
The Gouy module is available for download 22nd January 2010
and is featured at the
OpenWonderland Module Warehouse. This library is free software;
you can redistribute it and/or
modify it under the terms of the GNU Lesser General Public
License as published by the Free Software Foundation; either
version 2.1 of the License, or (at your option) any later version.
see the Wonderland Blog report
Symmetry operations in a 3D world
Many students indicate that the perception of molecular shapes in
3D causes real problems and the application of symmetry
operations is worse. By having molecular models with mirror
planes displayed in 3D where it is possible to view from all
angles or walk through or over etc may help to alleviate some of
these issues.
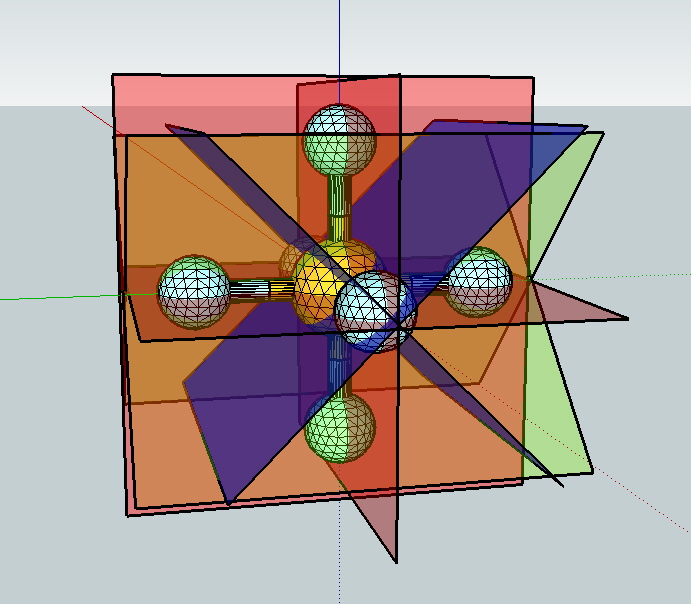
(A Sketchup model of SF
6)
Movie clips
We have prepared some short movie clips featuring a
spectroscopy laboratory and a GC trace obtained from pimento
berries. It shows the main features of the chromatograph as well
as models for some of the volatile components found in pimento
berries.
The files are LARGE > (10-20) MB so be prepared to wait for the
transfer...
Spectroscopy Laboratory
GCMS instrument
GC trace
H NMR of vinylpyrrolidinone
Raman spectrum of vanillin
Other models created by Craig and freely available from the Sketchup 3Dwarehouse
include this GC.
Acknowledgements
We are indebted to the Project Team for help throughout in our attempts to
deploy Wonderland on our Solaris servers.
The instrument models were generated by Craig Walters and Dominique Lyew.
© 2009-2018 by Robert John Lancashire, all
rights reserved.
Created and maintained by Prof. Robert J.
Lancashire,
The Department of Chemistry, University of the West Indies,
Mona Campus, Kingston 7, Jamaica.
Created July 2009. Last modified 21st March
2018






